分享到
Discovery of Replicating Circular RNAs by RNA-Seq and Computational Algorithms
Recently, a cooperation between Institute of Plant Protection, CAAS and School of Life Sciences, University of Science and Technology of China has yielded a big progress in the field of circular RNA (circRNA). Researchers discovered two novel replicating circRNAs from grapevine and apple plants using the further improved computational algorithm developed by the team. The research results have been published online in PLoS Pathogens on December 11, 2014.
RNA, a basic biomacromolecule, not only is able to carry and transmit genetic information, but also plays a critical role in regulation of gene expression. In addition to the classic RNAs of tRNA, mRNA, and rRNA, cells contain a striking diversity of additional RNA types including miRNAs, siRNAs, piRNAs, lncRNAs and other noncoding RNAs (ncRNA). They accomplish a remarkable variety of biological functions, regulating gene expression at the levels of transcription, RNA processing, and translation. “The versatility of RNA functions seem limitless. The latest surprise comes from circular RNAs” (Kosik, K. S., Nature, 2013, 495: 322-324). Thousands of non-replicating circRNAs have been identified in various organisms. Interestingly, a few circRNAs have been shown to play regulatory roles. However, the functions of circRNAs are largely remain unknown. Viroids have been proven to be excellent biological models for studying ncRNAs and circRNAs. But the rate of discovery of the replicating circular RNAs-viroids is still very low.
In this study, a considerably improved computational algorithms was developed by adopting parallel programming in the C++ language. The use of the new algorithms led to the discovery of a new viroid from grapevine and a viroid-like circular RNA from apple trees. Moreover, a new program was developed for the discovery of distinct classes of circular RNAs, including those neither replicate nor induce in vivo accumulation of Dicer-dependent siRNAs. The application of the new algorithms would speed up the discovery of novel circular RNAs and expand the list of known host species that can be infected by viroids.
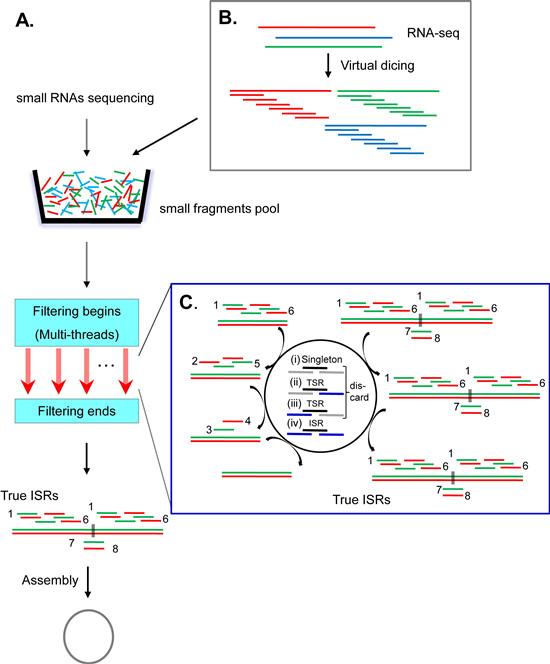
Illustration of the principles of the PFOR2 and SLS computational programs
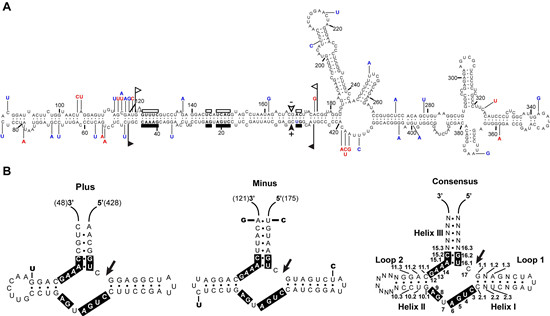
Primary and proposed secondary structures with minimum free energy for AHVd-like RNA
RNA, a basic biomacromolecule, not only is able to carry and transmit genetic information, but also plays a critical role in regulation of gene expression. In addition to the classic RNAs of tRNA, mRNA, and rRNA, cells contain a striking diversity of additional RNA types including miRNAs, siRNAs, piRNAs, lncRNAs and other noncoding RNAs (ncRNA). They accomplish a remarkable variety of biological functions, regulating gene expression at the levels of transcription, RNA processing, and translation. “The versatility of RNA functions seem limitless. The latest surprise comes from circular RNAs” (Kosik, K. S., Nature, 2013, 495: 322-324). Thousands of non-replicating circRNAs have been identified in various organisms. Interestingly, a few circRNAs have been shown to play regulatory roles. However, the functions of circRNAs are largely remain unknown. Viroids have been proven to be excellent biological models for studying ncRNAs and circRNAs. But the rate of discovery of the replicating circular RNAs-viroids is still very low.
In this study, a considerably improved computational algorithms was developed by adopting parallel programming in the C++ language. The use of the new algorithms led to the discovery of a new viroid from grapevine and a viroid-like circular RNA from apple trees. Moreover, a new program was developed for the discovery of distinct classes of circular RNAs, including those neither replicate nor induce in vivo accumulation of Dicer-dependent siRNAs. The application of the new algorithms would speed up the discovery of novel circular RNAs and expand the list of known host species that can be infected by viroids.

Illustration of the principles of the PFOR2 and SLS computational programs

Primary and proposed secondary structures with minimum free energy for AHVd-like RNA
More details are available on the bellow links:
http://www.plospathogens.org/article/info:doi/10.1371/journal.ppat.1004553
http://www.plospathogens.org/article/info:doi/10.1371/journal.ppat.1004553
Latest News
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
