Graph-based genome studies of Setaria — opening the doors of large-scale germplasms utilization and gene mining
Recently, Institute of Crop Sciences, CAAS (ICS-CAAS) has led an international research with scientists at New York University, USA, unlocking the graph-genome of Setaria , a C4 model plant. This significant advance propels the understanding of the domestication and improvement process of foxtail millet, the genetic basis for important agricultural traits, as well as the methodology for large-scale data-assisted plant breeding.
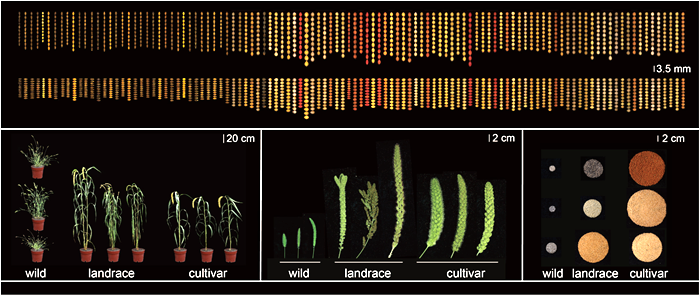
Researchers from ICS-CAAS systematically studied the evolution of foxtail millet, and de novo assembled 110 representative Setaria genomes selected from 1,844 varieties and accessions in world-wide germplasm collection, whose output has been published in Nature Genetics. They developed a graph-based genome and performed large-scale genetic studies for 68 traits across 22 environments in 13 geographical locations, identifying potential genes and marker-panels for foxtail millet improvement at different sites. According to the researchers, it serves as a significant milestone and will pave the way for the next generation of large-scale germplasm utilization and functional gene mining for future breeding. This publication narrows the gap for scientists to unlock the entire gene set of Setaria ( S. italica and S. viridis ). By understanding the comprehensive set of genomic variations, researchers will be equipped with superior genetic tools to pursue biological research and breeding of foxtail millet. Additionally, it provides an abundant resource for comparative genomics and “parallel domestication” studies with other crops.
This research has been highly praised by Prof. Li Jiayang and four other academicians from Chinese Academy of Sciences and Chinese Academy of Engineering: “The application of cutting-edge technologies in this research, such as high-quality graph-genomes, has enabled rapid and effective utilization of large-scale germplasm resources. Despite the challenges remain in exploring the mechanisms and utilization of detected causal genes or loci, the approach and methodologies adopted in this study hold substantial value as references for other crop research. They carry significant implications for enhancing the effective utilization of gene bank crop germplasm resources, which is crucial in ensuring national food security.”
The research is supported by grants from the National Key Research and Development Program of China (2021YFF1000100), the National Key R&D Program of China (2019YFD1000700/2019YFD1000701 and 2018YFD1000700), the National Natural Science Foundation of China (31871692 and 31871630), the China Agricultural Research System (CARS-06-13.5), the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, Strategic Priority Research Program of Chinese Academy of Sciences (grant XDPB16), the US National Science Foundation Plant Genome Research Program (IOS1546218 and 2204374) and the Zegar Family Foundation and the NYU Abu Dhabi Research Institute.
More details can be found at the link below: https://www.nature.com/articles/s41588-023-01423-w
By Diao Xianmin (diaoxianmin@caas.cn)
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
