分享到
IAS Constructed the CNV Map for Three Chinese Pig Breeds
Recently, the Collection, Conservation, Innovation and Utilization of Animal Resources team led by Professor Yuehui-Ma in Institute of Animal Science of CAAS, reported the identification and construction of the copy number variation (CNV) map for three Chinese native pig breeds.
CNV is a major form of structural genomic variation and has already been associated with diverse phenotypic variations in human as well as domesticated animals. Pig is an important domesticated animal and a significant large-animal model for medical research. However, the CNV coverage of the pig genome is still incomplete.
To extent the current coverage of CNVs in pig genome, the research team has performed a genome-wide CNV detection based on genotyping data of three previously unexamined Chinese pig breeds including Tibetan pig, Dahe pig and Wuzhishan pig. The result revealed a total of 105 CNV regions (CNVRs) and 25 (23.81%) among them were novel identified in this study compared with previous reports. The majority of these CNVRs overlap with known pig quantitative trait loci (QTLs) that affect meat quality, growth, reproduction and immune, suggesting a potential influence upon pig phenotypes of these CNVRs. Also, some CNVRs were found as harboring genes that had a human orthologous involved in human diseases such as cardiovascular and Alzheimer’s disease etc. This study provided valuables resources for follow-up associated studies of structural variations in pig complex traits as well as important implications of human health.
The results of the study have been published in Animal Genetics available here: http://onlinelibrary.wiley.com/doi/10.1111/age.12247/full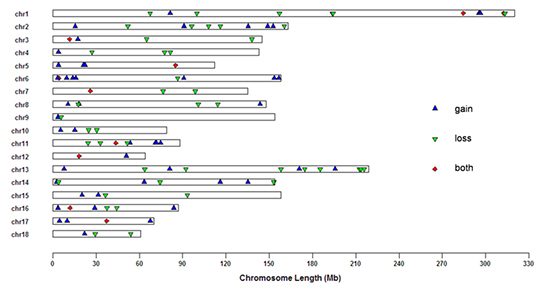
CNV is a major form of structural genomic variation and has already been associated with diverse phenotypic variations in human as well as domesticated animals. Pig is an important domesticated animal and a significant large-animal model for medical research. However, the CNV coverage of the pig genome is still incomplete.
To extent the current coverage of CNVs in pig genome, the research team has performed a genome-wide CNV detection based on genotyping data of three previously unexamined Chinese pig breeds including Tibetan pig, Dahe pig and Wuzhishan pig. The result revealed a total of 105 CNV regions (CNVRs) and 25 (23.81%) among them were novel identified in this study compared with previous reports. The majority of these CNVRs overlap with known pig quantitative trait loci (QTLs) that affect meat quality, growth, reproduction and immune, suggesting a potential influence upon pig phenotypes of these CNVRs. Also, some CNVRs were found as harboring genes that had a human orthologous involved in human diseases such as cardiovascular and Alzheimer’s disease etc. This study provided valuables resources for follow-up associated studies of structural variations in pig complex traits as well as important implications of human health.
The results of the study have been published in Animal Genetics available here: http://onlinelibrary.wiley.com/doi/10.1111/age.12247/full
By Dong Kunzhe
dkzhe1987@163.com
dkzhe1987@163.com

Latest News
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
