First report of a near-complete cucumber reference genome and multi-omics database
Recently, Cucurbitaceae Vegetable Genetics and Breeding Group in the Institute of Vegetables and Flowers (IVF, CAAS) made significant progress in cucumber genomics. The researchers accomplished the first-time near-complete assembly and gene annotation of the cucumber reference genome, along with the establishment of the inaugural comprehensive cucumber multi-omics database. The paper was published in Molecular Plant (IF=17.1) with the title of “A near-complete cucumber reference genome assembly and Cucumber-DB, a multi-omics database”.

Cucumber ( Cucumis sativus L.) is an important economic vegetable crop in the Cucurbitaceae family. Nearly 30% of the cucumber genome consists of complex repetitive sequences such as 45s rDNA and microsatellites, a proportion much higher than that of crops like rice, maize, and watermelon (< 5%). Due to limitations in sequencing technology and assembly methods at the time, the widely used reference genome of cucumber inbred line '9930' (CLv3.0 version) still has a large amount of unknown sequence (~130 Mb) and 72 gaps. Additionally, these repetitive sequences significantly impact the accuracy of gene annotation, highlighting the urgent need to improve the quality of the cucumber reference genome. Therefore, this study used the strategy of ONT+HiFi+HiC for the first time to obtain a near-complete assembly of the cucumber reference genome (CLv4.0) with only one remaining gap; based on large-scale PacBio full-length and Ilumina transcriptome data, a near-complete cucumber reference transcriptome dataset (CsRTD1) was constructed, with a BUSCO value of 99.19%. Integrating pan-genomic, population variation genomic, transcriptomic, and core germplasm information, the first cucumber multi-omics database, Cucumber-DB (http://www.cucumberdb.com/), was established, which can provide a comprehensive sharing platform for cucumber functional genomics and molecular breeding research.
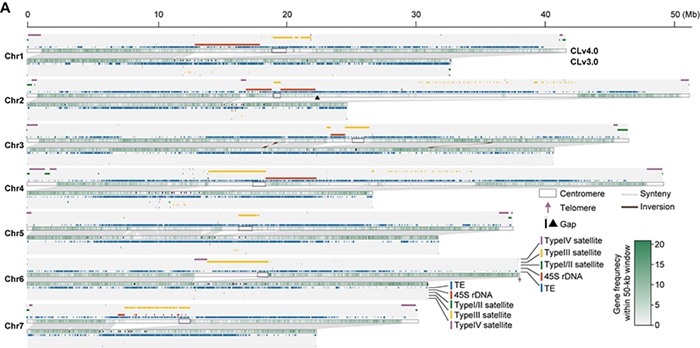
Figure 1 Comparison of Clv4.0 and Clv3.0 genome assemblies
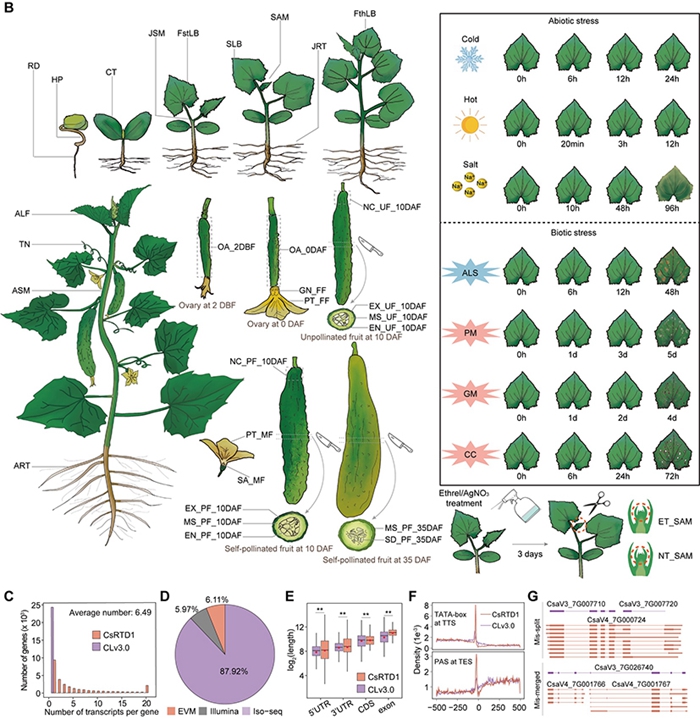
Figure 2 Construction and evaluation of the reference transcript dataset (CsRTD1)
This study was led by Prof. Zhang Shengping, as the co-corresponding author. Associate Prof. Guan Jiantao, Miao Han, Prof. Zhang Zhonghua, and Associate Prof. Dong Shaoyun contributed equally to this work as co-first authors. This work was funded by the National Key Research and Development Program, the Earmarked Fund for Modern Agro-industry Technology Research System, the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences, and the Central Public-interest Scientific Institution Basal Research Fund.

Figure 3 Overview of the cucumber multi-omics database (Cucumber-DB)
Link to this paper: https://www.cell.com/molecular-plant/fulltext/S1674-2052(24)00192-8
By Jiantao Guan (guanjiantao@caas.cn)
-
 Jul 03, 2024Experts from Italian National Agency for New Technologies, Energy and Sustainable Economic Development Visited IVF-CAAS
Jul 03, 2024Experts from Italian National Agency for New Technologies, Energy and Sustainable Economic Development Visited IVF-CAAS -
 Jun 28, 2024A cigar tobacco QX208, with excellent quality, disease and cold resistance, passed field evaluation
Jun 28, 2024A cigar tobacco QX208, with excellent quality, disease and cold resistance, passed field evaluation -
 Jun 18, 2024Visit by CGIAR Executive Director Ismahane Elouafi to the Institute of Vegetables and Flowers,CAAS
Jun 18, 2024Visit by CGIAR Executive Director Ismahane Elouafi to the Institute of Vegetables and Flowers,CAAS -
 May 29, 2024CAAS President Meets Deputy Director General of IAEA
May 29, 2024CAAS President Meets Deputy Director General of IAEA -
 May 29, 2024CAAS President Meets Chairman of the Understanding China Forum
May 29, 2024CAAS President Meets Chairman of the Understanding China Forum
