Peach Resource and Breeding Team Constructs First Comprehensive Peach Panvariomeand Develops Novel Strategy for High-Efficiency Gene Discovery
Recently, the peach resources and breeding group at the Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences (CAAS), announced a landmark achievement: the construction of the world's first comprehensive peach pan-variome atlas. This study identified 70.6% novel genomic variants and 3,289 previously unannotated genes, systematically elucidating global phylogenetic relationships and evolutionary trajectories of peach. The team also pioneered a panvariome-based genome-wide association study (GWASPV) strategy, enabling "one-step localization" of agronomic trait-associated genes and functional variants. This innovation significantly enhances gene discovery efficiency in woody fruit crops and provides robust theoretical and technical support for molecular breeding in peach. The findings were published as an article in the Molecular Plant .
Leveraging genome resequencing data from 1,020 peach germplasm accessions, the team systematically identified all known DNA variant types, including single nucleotide polymorphisms (SNPs), small insertions/deletions (indels), structural variations (SVs), copy number variations (CNVs), transposable element insertion polymorphisms (TIPs), and presence/absence variations (PAVs). The resulting pan-variome atlas revealed that 70.6% of variants were novel. Additionally, a pan-genome map of the 1,020 accessions uncovered 3,289 genes absent in the reference genome. Comparative analysis of gene counts across taxa demonstrated significant gene gains during peach domestication, contrasting with patterns observed in other plants and suggesting extensive gene introgression during peach evolution. Phylogenomic reconstruction traced global dissemination routes, revealing that almond contributed to the formation of wild P. davidiana , with P. mira identified as the most probable wild progenitor of cultivated peach. Furthermore, P. kansuensis was found to play a key role in the genetic architecture of P. ferganensis .
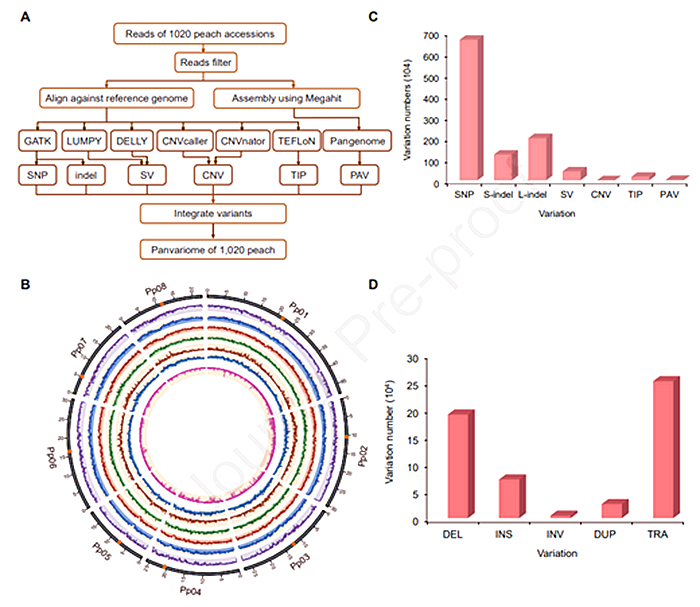
The study further revealed IBD segment sharing between wild and cultivated peaches, indicating a narrow genetic base and paucity of resistance genes in cultivated varieties. Integrated multi-omics analysis confirmed that most peach traits are polygenic, with a novel variant governing peach/nectarine was identified. The GWASPV strategy, integrating panvariome data, enabled high-throughput mining of over 2,000 trait-associated variants across 40 agronomic traits, 51 environmental adaptation traits, and 1,857 metabolite traits. This approach achieved "one-step resolution" of key genes and functional variants, markedly advancing gene discovery efficiency.
Dr. Yong Li, Associate Researcher at Zhengzhou Fruit Research Institute, served as the first author. Prof. Lirong Wang, Prof. Ke Cao, and Dr. Yong Li were corresponding authors. The research was supported by the National Key R&D Program of China (2023YFE0105400), the National Natural Science Foundation of China (32341042), the Chinese Academy of Agricultural Sciences Youth Project (Y2022QC23), the National Key Laboratory of Fruit and Vegetable Germplasm Innovation and Utilization, and the Zhongyuan New Wings Program Fund (ZYYWPF, ZYZX20240304). Additional funding was provided by the CAAS Innovation Project (CAAS-ASTIP-2024-ZFRI-01).
By liyong02@caas.cn
-
 Jun 05, 2025CAAS President Meets New FAO Representative in China
Jun 05, 2025CAAS President Meets New FAO Representative in China -
 May 27, 2025Director General of IARRP attends 2025 Global Forum of Group on Earth Observations
May 27, 2025Director General of IARRP attends 2025 Global Forum of Group on Earth Observations -
 May 20, 2025IPPCAAS Experts Visit Australia to Promote In-Depth China–Australia Cooperation in Plant Biosafety
May 20, 2025IPPCAAS Experts Visit Australia to Promote In-Depth China–Australia Cooperation in Plant Biosafety -
 May 20, 2025Opening Ceremony of the Seminar on Modern Breeding and Management of the Vegetable Industry for BRI Partner Countries Successfully Held in Yanqing, Beijing
May 20, 2025Opening Ceremony of the Seminar on Modern Breeding and Management of the Vegetable Industry for BRI Partner Countries Successfully Held in Yanqing, Beijing -
 May 18, 2025Sustainable Dairy Development — The 9th International Symposium on Dairy Cow Nutrition and Milk Quality Convenes in Beijing
May 18, 2025Sustainable Dairy Development — The 9th International Symposium on Dairy Cow Nutrition and Milk Quality Convenes in Beijing
