T2T Genome Assemblies of Pigs Reveal Genes Associated with Cold and Heat Tolerance
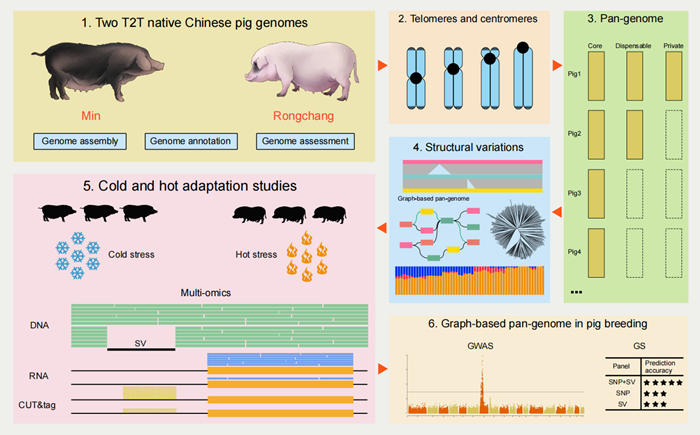
Recently, the Swine Genetics and Breeding Innovation Team at the Institute of Animal Science, Chinese Academy of Agricultural Sciences (CAAS), systematically revealed the significant value and broad application prospects of telomere-to-telomere (T2T) genome research in pigs. Their study encompassed T2T genome assembly, centromeric characteristics, structural variation (SV) detection using third-generation sequencing, construction of a graph-based pangenome, and the identification of genes associated with cold and heat tolerance. The related findings were published in iMeta .
Currently, the international reference genome for pigs is based on the Duroc breed and serves as the cornerstone for pig genomics research. However, this reference has not yet reached the T2T level, which limits the advancement of pig genomics. China spans climates from frigid to tropical regions and possesses the richest pig breed diversity in the world.
Among them, Min pigs from the cold Northeastern region and Rongchang pigs from the humid and hot Southwestern area are representative breeds known for their strong cold and heat tolerance, respectively. This study successfully assembled T2T genomes of both Min and Rongchang pigs and provided novel insights into the structural features of telomeric and centromeric regions.
Through pangenome analysis, researchers identified 25,472 gene families, of which 38.94% were core gene families, 60.09% variable gene families, and 0.97% private gene families. In addition, they identified 194,234 high-quality structural variants and constructed a graph-based pangenome. By integrating multi-omics data, the team discovered the key gene TPT1 related to cold tolerance in Min pigs and GIMAP6 related to heat tolerance in Rongchang pigs. A potentially critical mutation upstream of GIMAP6 was also identified, which may enhance the heat tolerance of Rongchang pigs. Furthermore, the construction of the graph-based pangenome significantly improved the detection of quantitative trait loci (QTLs) in genome-wide association studies (GWAS) and enhanced the accuracy of genomic selection (GS) breeding.
These findings provide both theoretical foundations and technical support for future studies on climate adaptability in pigs and related breeding programs in the context of global warming.
This research was supported by the National Key R&D Program of China, The Strategic Cooperation Funding Project between People's Government of Chongqing Municipality and CAAS, The design and cultivation of the new breed of pigs with high-quality meat, and the Science and Technology Innovation Program of CAAS.
The full article can be accessed through the Wiley Online Library at the following link: https://onlinelibrary.wiley.com/doi/10.1002/imt2.70013
-
 Apr 25, 2025CAAS President Meets Director General of IAEA
Apr 25, 2025CAAS President Meets Director General of IAEA -
 Apr 25, 2025CAAS Delegation Attends CGIAR Science Week 2025
Apr 25, 2025CAAS Delegation Attends CGIAR Science Week 2025 -
 Apr 25, 2025Delegation Led by CEO of Punjab Agricultural Research Council Abid Mahmood and Vice Dean of PKU Advanced Agricultural Research Institute Zhang Xingping Visits Shouguang R&D Center of CAAS
Apr 25, 2025Delegation Led by CEO of Punjab Agricultural Research Council Abid Mahmood and Vice Dean of PKU Advanced Agricultural Research Institute Zhang Xingping Visits Shouguang R&D Center of CAAS -
 Apr 11, 2025Strengthening China-Mongolia Grassland Cooperation to Build a Cross-Border Green Ecological Barrier
Apr 11, 2025Strengthening China-Mongolia Grassland Cooperation to Build a Cross-Border Green Ecological Barrier -
 Apr 03, 2025CAAS and CABI Forge a New Chapter in Strategic Cooperation
Apr 03, 2025CAAS and CABI Forge a New Chapter in Strategic Cooperation
