Structural Variation Research Drives a New Breakthrough in Alfalfa Genetic Improvement
Recently, the Forage Breeding and Cultivation Technology Innovation Team at the Institute of Animal Science, Chinese Academy of Agricultural Sciences (CAAS), successfully constructed a high-quality pan-genome for alfalfa and systematically elucidated its genetic diversity, as well as the genetic basis underlying salt tolerance and quality-related traits. This work represents a significant advancement in alfalfa genomics, propelling functional gene discovery and molecular breeding into a new phase. The findings were published in Nature Genetics .
Alfalfa, widely known as the "King of Forage," is a high-quality feed for ruminant livestock such as cattle and sheep due to its high protein content, abundant forage yield, and richness in vitamins, minerals, and other bioactive compounds. In addition to its nutritional value, alfalfa enhances soil fertility through nitrogen fixation by root nodules, thereby reducing the need for chemical fertilizers and significantly improving the yield of subsequent crops. Notably, cultivation of alfalfa in saline-alkali soils can markedly reduce soil salinity and alkalinity. However, the species' highly complex autotetraploid genome, characterized by high levels of heterozygosity and abundant structural variations, poses major challenges for the precise identification and effective utilization of genes associated with important agronomic traits. Although several alfalfa genome assemblies have been released in recent years owing to advances in sequencing and assembly technologies, relying solely on a single reference genome is insufficient to unravel the genetic basis underlying traits such as yield, quality, and stress resistance. This limitation severely hampers accurate gene mapping and the genetic improvement of alfalfa.
To address these challenges, the research team constructed a high-quality pangenome by integrating genomic data from 24 alfalfa accessions with diverse genetic backgrounds. Based on this resource, we performed structural variation-based genome-wide association studies (SV-GWAS) and identified key structural variants associated with salt tolerance and forage quality. In-depth analysis of candidate genes uncovered MsMAP65 , a gene involved in salt stress regulation, and MsGA3ox1 , a critical regulator of stem and leaf development. Furthermore, genomic selection revealed numerous genetic markers with significant breeding potential, providing both theoretical insights and practical strategies for the coordinated improvement of key agronomic traits in alfalfa.
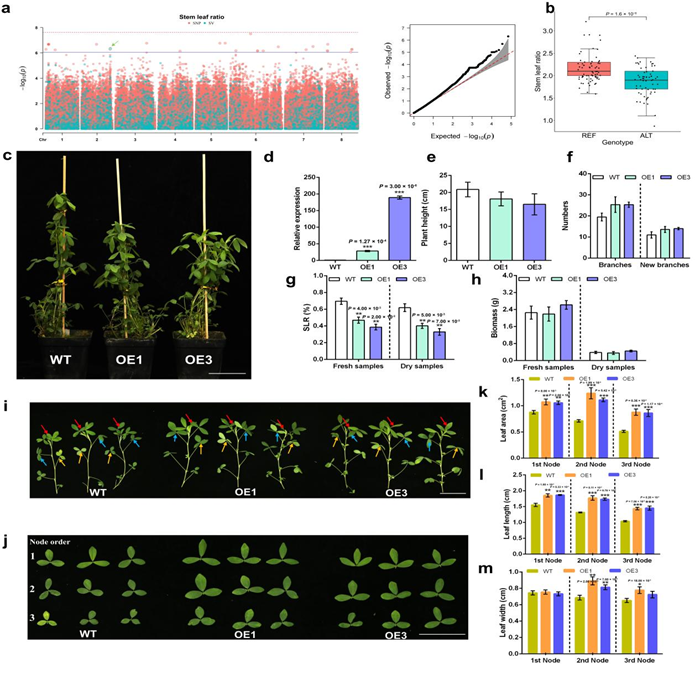
Functional validation of a key gene identified by pan-genomic analysis of SLR phenotype using SNP-GWAS and SV-GWAS
This work was supported by the National Forage Industry Technology System, the Agricultural Biological Breeding Project, the Key Projects in Science and Technology of Inner Mongolia, and the Science and Technology Innovation Program of CAAS.
DOI: https://www.nature.com/articles/s41588-025-02164-8
-
 Apr 25, 2025CAAS President Meets Director General of IAEA
Apr 25, 2025CAAS President Meets Director General of IAEA -
 Apr 25, 2025CAAS Delegation Attends CGIAR Science Week 2025
Apr 25, 2025CAAS Delegation Attends CGIAR Science Week 2025 -
 Apr 25, 2025Delegation Led by CEO of Punjab Agricultural Research Council Abid Mahmood and Vice Dean of PKU Advanced Agricultural Research Institute Zhang Xingping Visits Shouguang R&D Center of CAAS
Apr 25, 2025Delegation Led by CEO of Punjab Agricultural Research Council Abid Mahmood and Vice Dean of PKU Advanced Agricultural Research Institute Zhang Xingping Visits Shouguang R&D Center of CAAS -
 Apr 11, 2025Strengthening China-Mongolia Grassland Cooperation to Build a Cross-Border Green Ecological Barrier
Apr 11, 2025Strengthening China-Mongolia Grassland Cooperation to Build a Cross-Border Green Ecological Barrier -
 Apr 03, 2025CAAS and CABI Forge a New Chapter in Strategic Cooperation
Apr 03, 2025CAAS and CABI Forge a New Chapter in Strategic Cooperation
